Plot Occurrence Trends or Range Size Trends for Individual Species
Source:R/plot_species_ts.R
plot_species_ts.RdCreates time series plots of species occurrences or species
range sizes, with an optional smoothed trendline, and visualizes
uncertainty. Requires an indicator_ts object created using the
spec_occ_ts() or spec_range_ts() functions as input. To plot
multi-species indicators (e.g., species richness or evenness), use the
plot_ts() function instead.
Usage
plot_species_ts(
x,
species,
single_plot = TRUE,
min_year = NULL,
max_year = NULL,
title = "auto",
auto_title = NULL,
y_label_default = NULL,
suppress_y = FALSE,
smoothed_trend = TRUE,
linecolour = NULL,
linealpha = 0.8,
ribboncolour = NULL,
ribbonalpha = 0.2,
error_alpha = 1,
trendlinecolour = NULL,
trendlinealpha = 0.5,
envelopecolour = NULL,
envelopealpha = 0.2,
smooth_cialpha = 1,
point_line = c("point", "line"),
pointsize = 2,
linewidth = 1,
ci_type = c("error_bars", "ribbon"),
error_width = 1,
error_thickness = 1,
smooth_linetype = c("solid", "dashed", "dotted", "dotdash", "longdash", "twodash"),
smooth_linewidth = 1,
smooth_cilinewidth = 1,
gridoff = FALSE,
x_label = NULL,
y_label = NULL,
x_expand = 0.1,
y_expand = 0.1,
x_breaks = 10,
y_breaks = 6,
title_wrap_length = 60,
spec_name_wrap_length = 40
)Arguments
- x
An 'indicator_ts' object containing time series of indicator values matched to species names and/or taxon keys, created using the
spec_occ_ts()orspec_range_ts()functions. This is a required parameter with no default.- species
Species you want to map occurrences for. Can be either numerical taxonKeys or species names. Partial species names can be used (the function will try to match them). This is a required parameter with no default.
- single_plot
(Optional) By default all species occurrence time series will be combined into a single multi-panel plot. Set this to FALSE to plot each species separately.
- min_year
(Optional) Earliest year to include in the plot.
- max_year
(Optional) Latest year to include in the plot.
- title
(Optional) Plot title. Replace "auto" with your own title if you want a custom title or if calling the function manually.
- auto_title
(Optional) Text for automatic title generation, provided by an appropriate S3 method (if calling the function manually, leave as NULL).
- y_label_default
(Optional) Default label for the y-axis, provided by an appropriate S3 method (if calling the function manually, leave as NULL).
- suppress_y
(Optional) If TRUE, suppresses y-axis labels.
- smoothed_trend
(Optional) If TRUE, plot a smoothed trendline over time (
stats::loess()).- linecolour
(Optional) Colour for the indicator line or points. Default is darkorange.
- linealpha
(Optional) Transparency for indicator line or points. Default is 0.8.
- ribboncolour
(Optional) Colour for the bootstrapped confidence intervals. Default is goldenrod1. Set to "NA" if you don't want to plot the CIs.
- ribbonalpha
(Optional) Transparency for indicator confidence interval ribbon (if ci_type = "ribbon"). Default is 0.2.
- error_alpha
(Optional) Transparency for indicator error bars (if ci_type = "error_bar"). Default is 1.
- trendlinecolour
(Optional) Colour for the smoothed trendline. Default is blue.
- trendlinealpha
(Optional) Transparency for the smoothed trendline. Default is 0.5.
- envelopecolour
(Optional) Colour for the uncertainty envelope. Default is lightsteelblue.
- envelopealpha
(Optional) Transparency for the smoothed trendline envelope. Default is 0.2.
- smooth_cialpha
(Optional) Transparency for the smoothed lines forming the edges of the trendline envelope. Default is 1.
- point_line
(Optional) Whether to plot the indicator as a line or a series of points. Options are "line" or "point". Default is "point".
- pointsize
(Optional) Size of the points if point_line = "point". Default is 2.
- linewidth
(Optional) Width of the line if point_line = "line". Default is 1.
- ci_type
(Optional) Whether to plot bootstrapped confidence intervals as a "ribbon" or "error_bars". Default is "error_bars".
- error_width
(Optional) Width of error bars if ci_type = "error_bars". Default is 1. Note that unlike the default 'width' parameter in geom_errorbar, 'error_width' is NOT dependent on the number of data points in the plot. It is automatically scaled to account for this. Therefore the width you select will be consistent relative to the plot width even if you change 'min_year' and 'max_year'.
- error_thickness
(Optional) Thickness of error bars if ci_type = "error_bars". Default is 1.
- smooth_linetype
(Optional) Type of line to plot for smoothed trendline. Default is "solid".
- smooth_linewidth
(Optional) Line width for smoothed trendline. Default is 1.
- smooth_cilinewidth
(Optional) Line width for smoothed trendline confidence intervals. Default is 1.
- gridoff
(Optional) If TRUE, hides gridlines.
- x_label
(Optional) Label for the x-axis.
- y_label
(Optional) Label for the y-axis.
- x_expand
(Optional) Expansion factor to expand the x-axis beyond the data. Left and right values are required in the form of c(0.1, 0.2) or simply 0.1 to apply the same value to each side. Default is 0.05.
- y_expand
(Optional) Expansion factor to expand the y-axis beyond the data. Lower and upper values are required in the form of c(0.1, 0.2) or simply 0.1 to apply the same value to the top and bottom. Default is 0.05.
- x_breaks
(Optional) Integer giving desired number of breaks for x axis. (May not return exactly the number requested.)
- y_breaks
(Optional) Integer giving desired number of breaks for y axis. (May not return exactly the number requested.)
- title_wrap_length
(Optional) Maximum title length before wrapping to a new line.
- spec_name_wrap_length
(Optional) Maximum species name length before wrapping to a new line.
Value
A ggplot object representing species range or occurrence time series plot(s). Can be customized using ggplot2 functions.
Examples
spec_occ_ts_mammals_denmark <- spec_occ_ts(example_cube_1,
level = "country",
region = "Denmark")
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 2 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 2 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 22 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 2 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 3 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
#> [1] "All values of t are equal to 1 \n Cannot calculate confidence intervals"
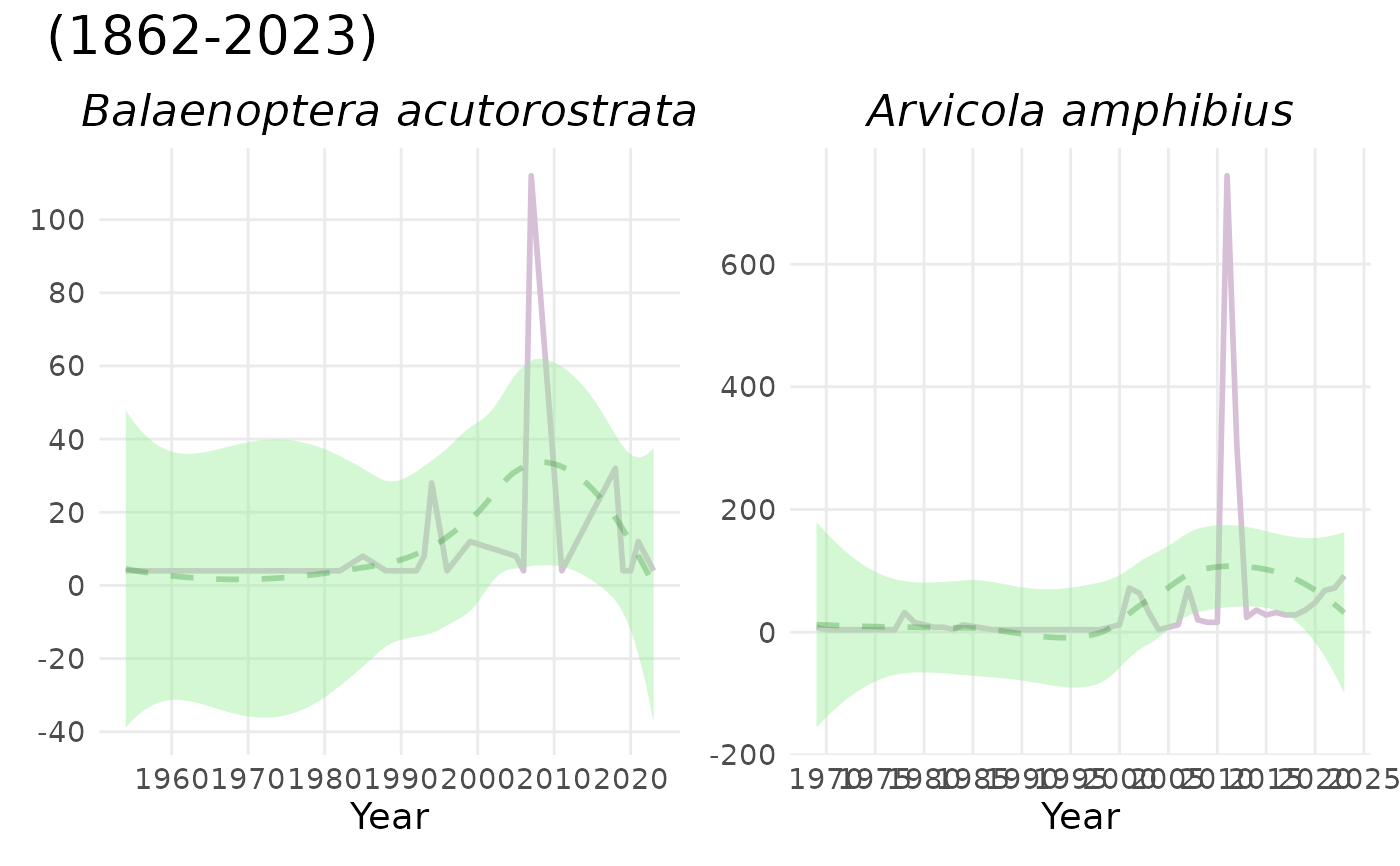
# default colours:
plot_species_ts(spec_occ_ts_mammals_denmark, c(2440728, 4265185))
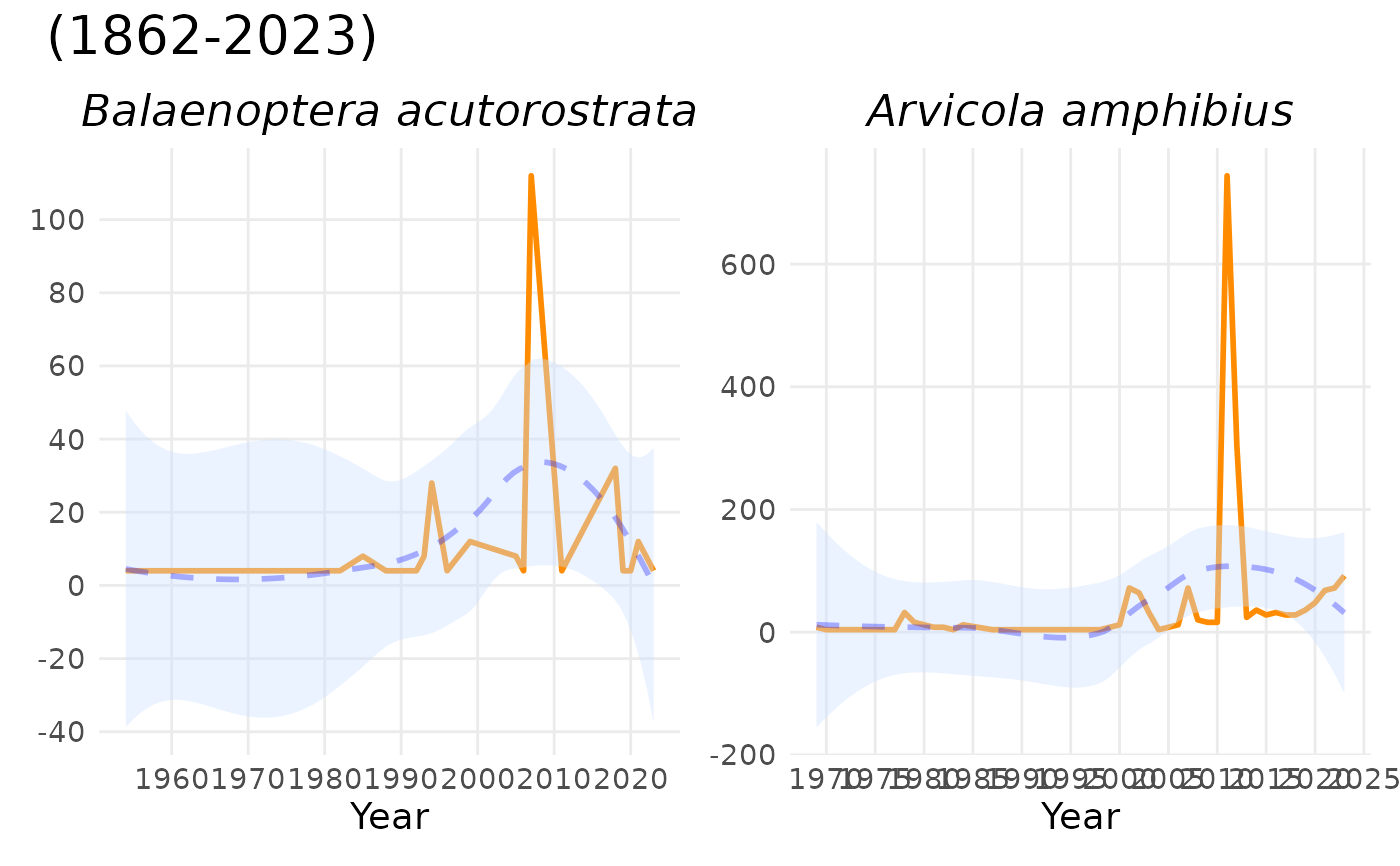 # custom colours:
plot_species_ts(spec_occ_ts_mammals_denmark, c(2440728, 4265185),
linecolour = "thistle",
trendlinecolour = "forestgreen",
envelopecolour = "lightgreen")
# custom colours:
plot_species_ts(spec_occ_ts_mammals_denmark, c(2440728, 4265185),
linecolour = "thistle",
trendlinecolour = "forestgreen",
envelopecolour = "lightgreen")